We are recruiting motivated students and postdoctoral fellows.
Please get in touch if you are interested in joining our team.
We support diversity and inclusion and flexible working.
Our lab is at the interface of epigenetics and immunology and we seek to understand how ‘dark matter’, which is DNA of unknown function, can contribute to normal immune regulation as well as inflammation in cancer and autoimmunity.
What is dark matter?
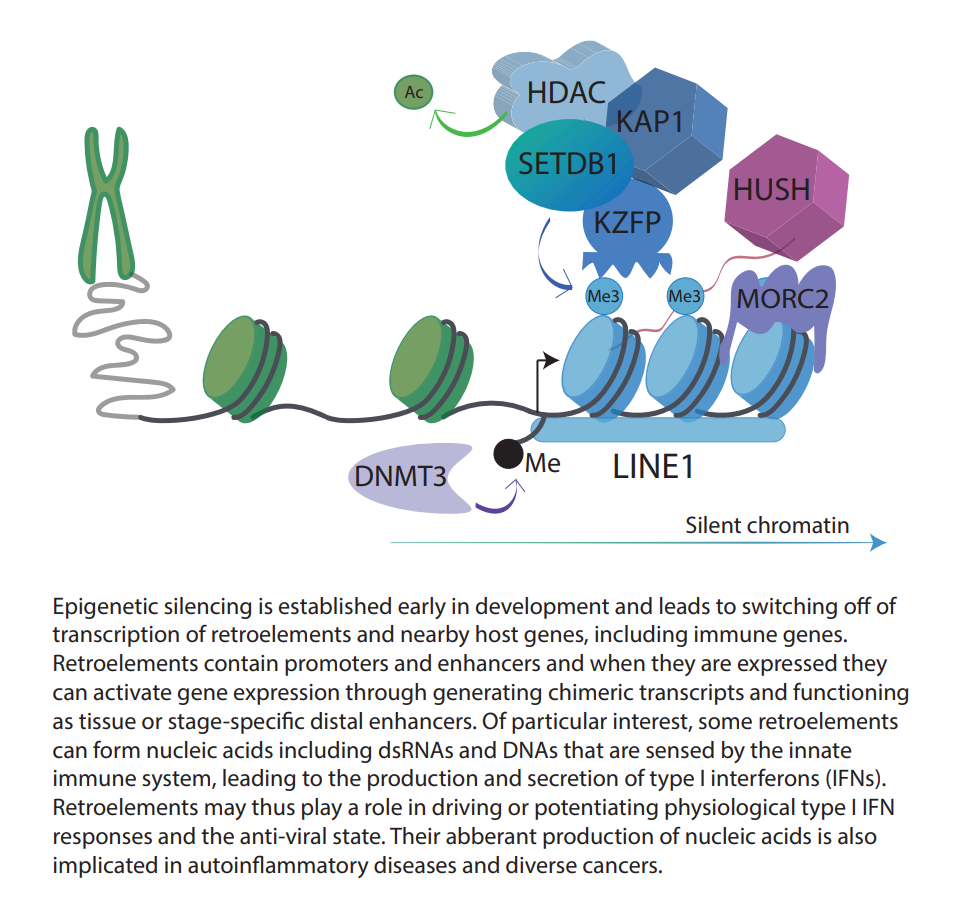
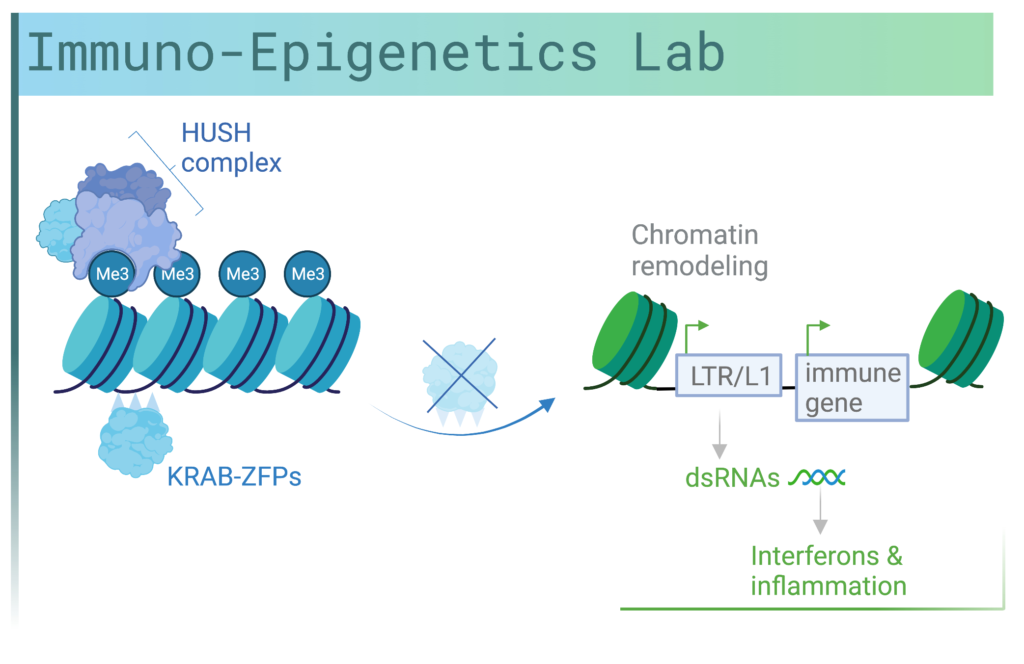
Retroelements form part of the ‘dark matter’ within our genome, due to their poorly characterized functions. Once ancient viruses that invaded our genome, these elements can still retain enhancers and promoters that regulate our own genes, with rare copies of them still able to replicate. A critical layer of regulation of retroelements is through epigenetic modifications, including the heterochromatin-associated histone mark, histone 3 lysine 9 trimethylation (H3K9me3). Retroelements are linked to inflammation through two molecular mechanisms: firstly, they can switch ON and OFF immune genes directly, and secondly, they can generate nucleic acids that the immune system detects as foreign through RNA and DNA sensors leading to a type I interferon response. Type I interferon signaling activates interferon response genes with diverse roles in inflammation.
Why is this important?
We study the cross-talk between chromatin regulation and inflammation in human adult tissues. Acute Inflammation is part of the healthy immune response, which acts to clear damaged or infected cells. In contrast, inflammation that is not resolved (chronic) is a hallmark of cancer and autoimmunity. Autoimmunity ensues when our white blood cells attack our own body. In cancer, Inflammation promotes immune evasion through driving immunosuppressive cell infiltration and tumour invasiveness. Downregulation of chromatin-associated factors leads to remodeling of chromatin state through epigenetic modifications (histone acetylation and methylation and DNA methylation). In our lab, we are interested in how chromatin remodeling and epigenetic memory can drive cancer progression and autoimmune disease through inflammation. Understanding these pathways will lead to innovative therapies for cancer and autoimmune diseases.
Our research
The human immune system encompasses innate and adaptive (antigen-specific) branches and defends us from pathogens and damage through inflammation. Intriguingly, mounting evidence from our lab and other labs has shown that LINE-1 and related retroelements can induce type I interferon and NFkB signaling pathways, which are central to the activation of the innate and adaptive immune system. This is akin to viruses inducing interferons through nucleic acid sensing of their viral replication intermediates. Type I interferons are secreted and render bystander cells in an anti-viral state and they are also fundamental to the recruitment of T cells and B cells to clear pathogens and to drive anti-tumour immunity. Persistent type I interferon signalling, in contrast, is a hallmark of inflammatory and autoimmune diseases such as systemic lupus erythematosus (SLE).
Recent evidence from our lab and other labs suggests that a key layer of regulation of inflammation is at the chromatin level through epigenetic mechanisms (DNA methylation, histone modifications and noncoding RNAs). In our lab, we study the chromatin regulation of immune genes in human adult tissues and how dysregulation of epigenetic pathways can contribute to cancer and inflammatory diseases. Loss of epigenetic regulators can occur following environmental challenges that lead to mutations in genes encoding these factors or in genes that regulate them. We focus on transcription factors known as KRAB-zinc finger proteins (KRAB-ZFPs) and the epigenetic regulators, TRIM28 and the HUSH (human silencing hub) complex. Surprisingly, we have discovered these factors to bind to cis-regulatory elements derived from retroelements. Targeted disruption of these transcriptional regulators leads to changes in immune gene expression through remodeling of chromatin.
Our research on the chromatin control of inflammation will pave the way for new cancer therapies based on awakening anti-tumour immune responses and for novel strategies to tackle the underlying causes of inflammation more broadly.
Current lab projects
- HUSH (human silencing hub) complex regulation of inflammation
- KRAB-zinc finger protein control of gene-regulatory networks